Palmerpenquins tidymodels
By Edward F. Hillenaar
January 1, 0001
Get started with tidymodels and #TidyTuesday Palmer penguins
This is a blogpost of Julia Silge about rstats tidymodels: Julia Silge’s youtube video.
Palmerpenguins dataset
The Palmerpenguins dataset can be found here:
Palmerpenguins dataset.
We can build a classification model to distinguish male and female penguins.
Explore data
glimpse(penguins)
## Rows: 344
## Columns: 8
## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
## $ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
## $ sex <fct> male, female, female, NA, female, male, female, male…
## $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…
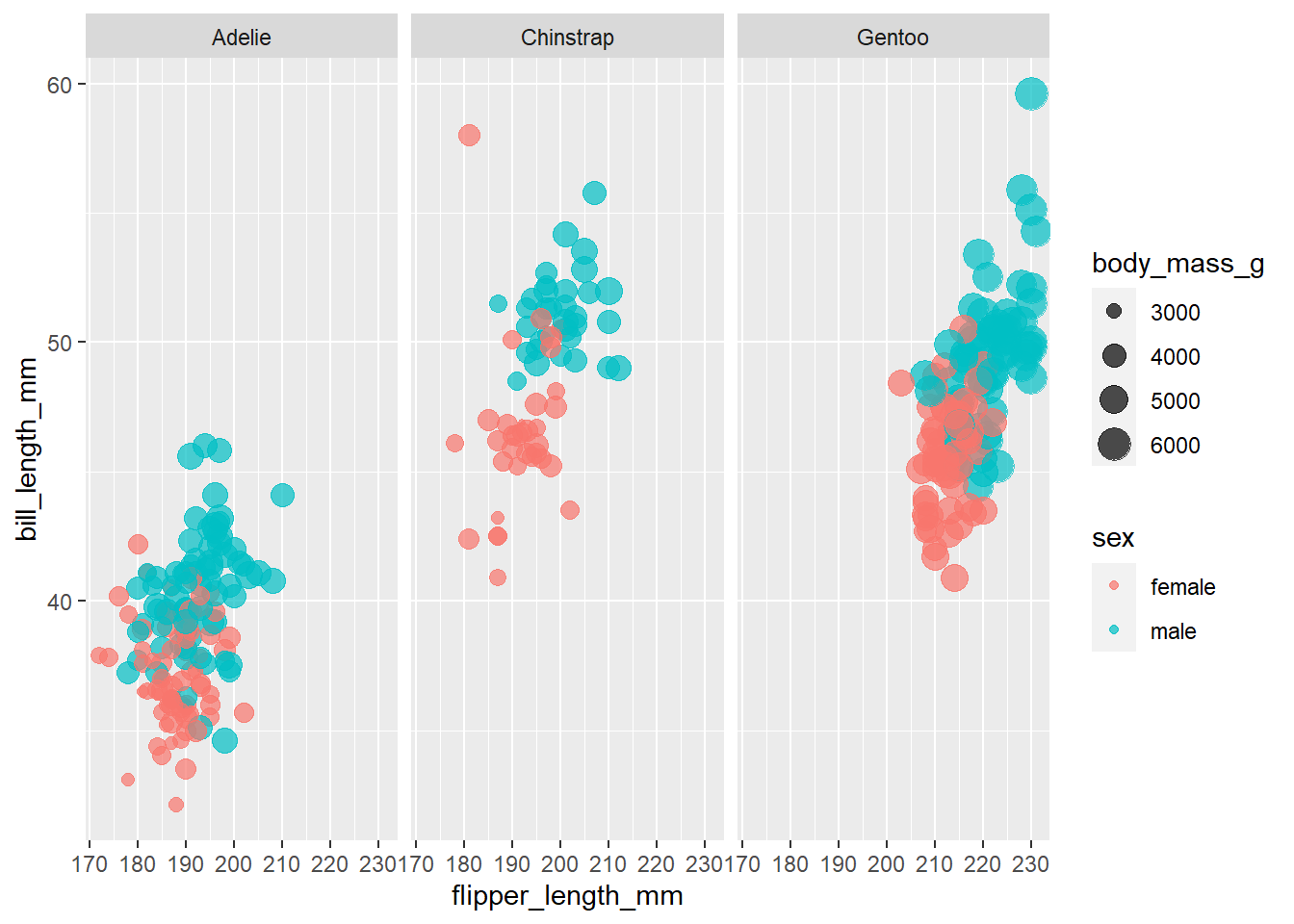
penguins %>%
filter(!is.na(sex)) %>%
ggplot(aes(flipper_length_mm, bill_length_mm, color = sex, size = body_mass_g)) +
geom_point(alpha = 0.7) +
facet_wrap(~species)
penguins_df <- penguins %>%
filter(!is.na(sex)) %>%
select(-year, -island)
Build a model
set.seed(123)
penguin_split <- initial_split(penguins_df, strata = sex)
penguin_train <- training(penguin_split)
penguin_test <- testing(penguin_split)
library(rsample)
# Set seed for reproducibility
set.seed(123)
# Ungroup the data if it's grouped
penguin_train <- ungroup(penguin_train)
# Number of bootstrap replicates
num_replicates <- 250
# Create an empty list to store bootstrap samples
penguin_boot <- vector("list", num_replicates)
# Perform bootstrap resampling
for (i in 1:num_replicates) {
# Generate random indices with replacement
indices <- sample(nrow(penguin_train), replace = TRUE)
# Extract bootstrap sample using the random indices
penguin_boot[[i]] <- penguin_train[indices, ]
}
# View the first bootstrap sample
penguin_boot[[1]]
## # A tibble: 249 × 6
## species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex
## <fct> <dbl> <dbl> <int> <int> <fct>
## 1 Adelie 38.3 19.2 189 3950 male
## 2 Gentoo 48.6 16 230 5800 male
## 3 Adelie 41.5 18.5 201 4000 male
## 4 Adelie 37 16.9 185 3000 female
## 5 Gentoo 49.6 16 225 5700 male
## 6 Adelie 37.7 19.8 198 3500 male
## 7 Adelie 32.1 15.5 188 3050 female
## 8 Chinstrap 42.5 17.3 187 3350 female
## 9 Adelie 38.8 17.6 191 3275 female
## 10 Chinstrap 51.3 18.2 197 3750 male
## # ℹ 239 more rows
Setting up a (regression) model to train the data
glm_spec <- logistic_reg() %>%
set_engine("glm")
rf_spec <- rand_forest() %>%
set_mode("classification") %>%
set_engine("ranger")
penguin_wf <- workflow() %>%
add_formula(sex ~ .)
penguin_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: None
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## sex ~ .
Evaluate modeling
collect_metrics(rf_rs)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.919 25 0.0164 Preprocessor1_Model1
## 2 roc_auc binary 0.977 25 0.00807 Preprocessor1_Model1
collect_metrics(glm_rs)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.919 25 0.0174 Preprocessor1_Model1
## 2 roc_auc binary 0.981 25 0.00770 Preprocessor1_Model1
glm_rs %>%
conf_mat_resampled()
## # A tibble: 4 × 3
## Prediction Truth Freq
## <fct> <fct> <dbl>
## 1 female female 4.52
## 2 female male 0.4
## 3 male female 0.4
## 4 male male 4.64
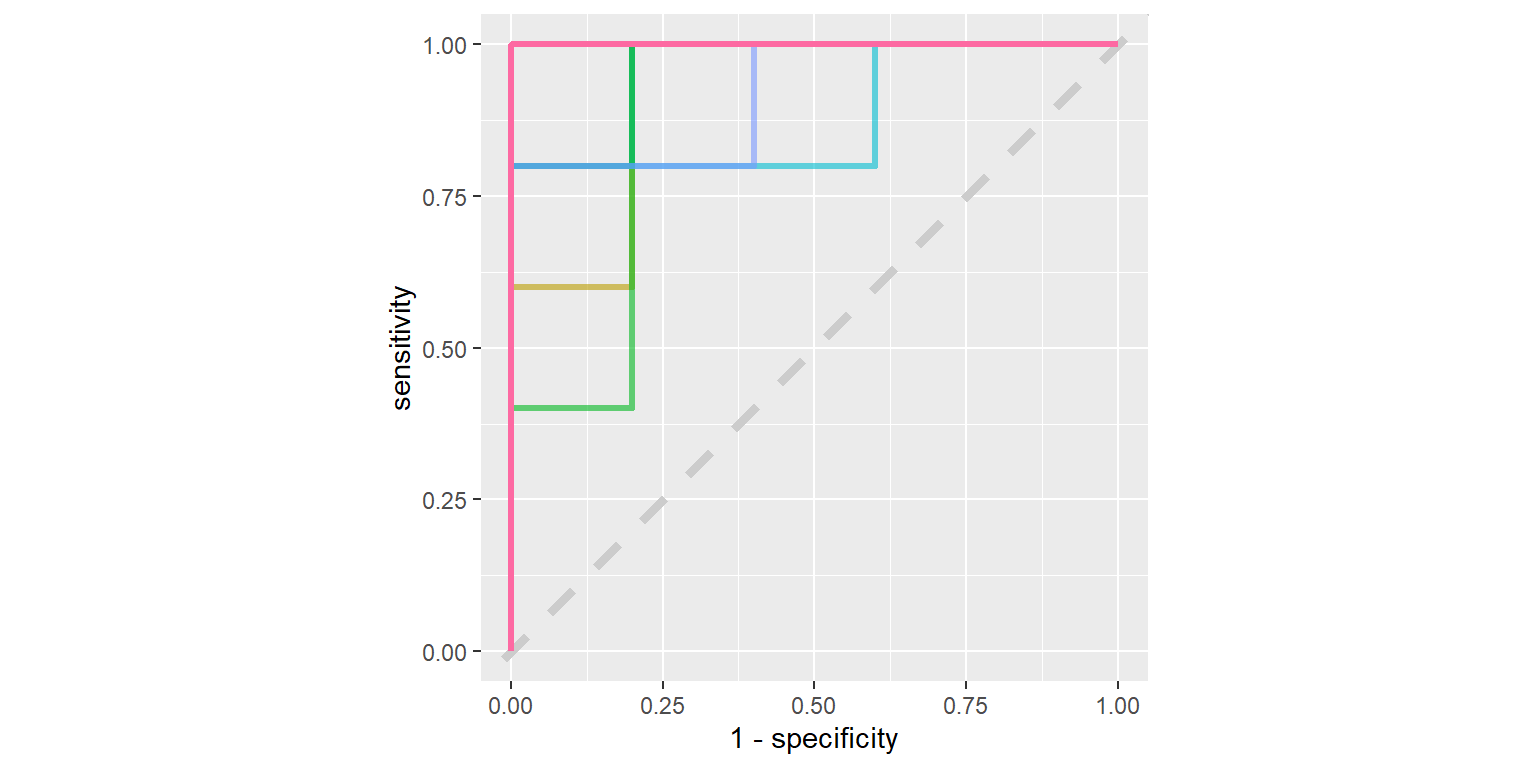
glm_rs %>%
collect_predictions() %>%
group_by(id) %>%
roc_curve(sex, .pred_female) %>%
ggplot(aes(1 - specificity, sensitivity, color = id)) +
geom_abline(lty = 2, color = "gray80", size = 1.5) +
geom_path(show.legend = FALSE, alpha = 0.6, size = 1.2) +
coord_equal()
## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
penguin_final <- penguin_wf %>%
add_model(glm_spec) %>%
last_fit(penguin_split)
penguin_final
## # Resampling results
## # Manual resampling
## # A tibble: 1 × 6
## splits id .metrics .notes .predictions .workflow
## <list> <chr> <list> <list> <list> <list>
## 1 <split [249/84]> train/test split <tibble> <tibble> <tibble> <workflow>
collect_metrics(penguin_final)
## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.857 Preprocessor1_Model1
## 2 roc_auc binary 0.938 Preprocessor1_Model1
collect_predictions(penguin_final) %>% conf_mat(sex, .pred_class)
## Truth
## Prediction female male
## female 37 7
## male 5 35
penguin_final$.workflow[[1]] %>%
tidy(exponentiate = TRUE) %>% arrange(estimate)
## # A tibble: 7 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 5.75e-46 19.6 -5.31 0.000000110
## 2 speciesGentoo 1.14e- 5 3.75 -3.03 0.00243
## 3 speciesChinstrap 1.37e- 4 2.34 -3.79 0.000148
## 4 body_mass_g 1.01e+ 0 0.00176 4.59 0.00000442
## 5 flipper_length_mm 1.06e+ 0 0.0611 0.926 0.355
## 6 bill_length_mm 1.91e+ 0 0.180 3.60 0.000321
## 7 bill_depth_mm 8.36e+ 0 0.478 4.45 0.00000868
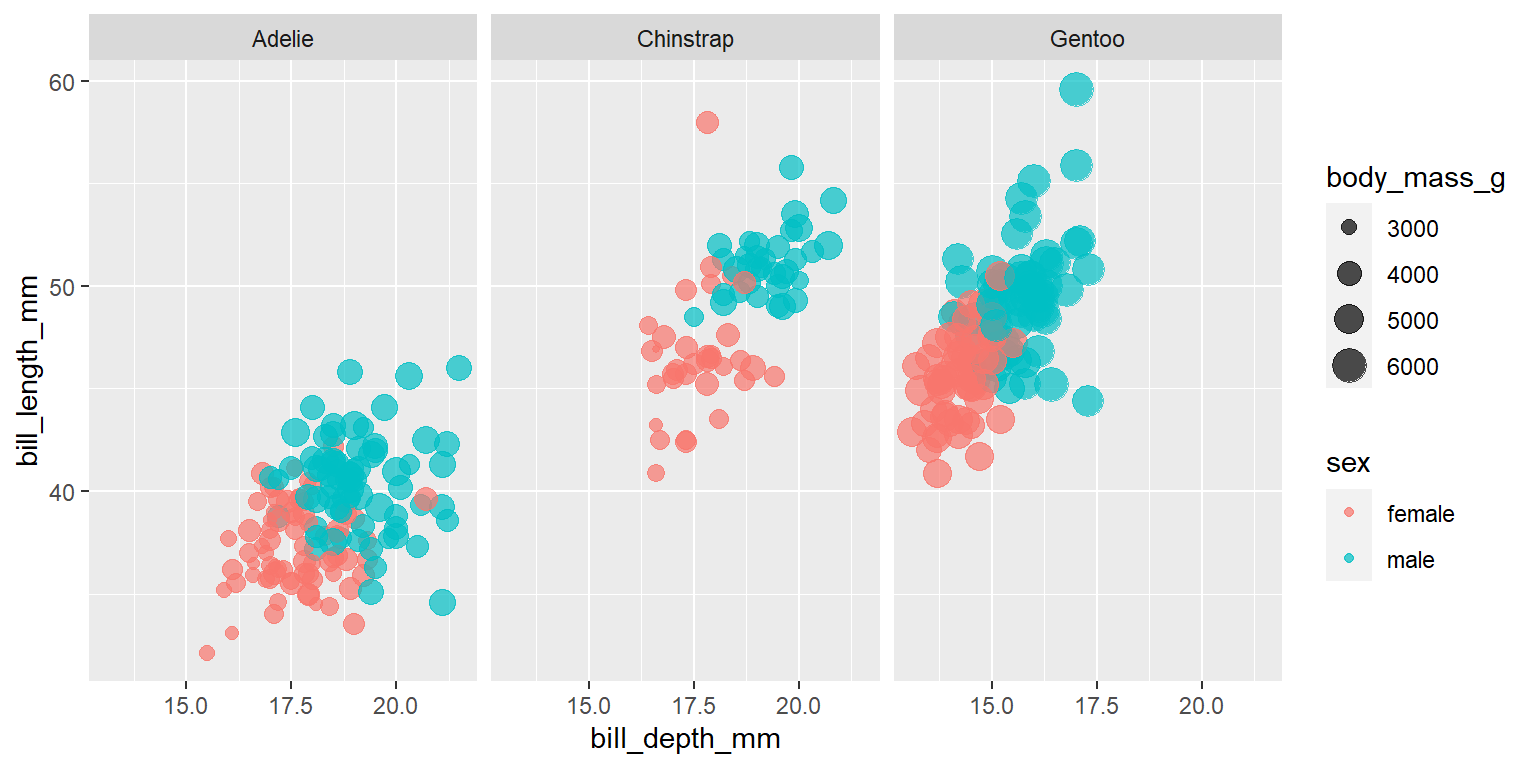
penguins %>%
filter(!is.na(sex)) %>%
ggplot(aes(bill_depth_mm, bill_length_mm, color = sex, size = body_mass_g)) +
geom_point(alpha = 0.7) +
facet_wrap(~species)
References
Gorman, K. B., Williams, T. D., & Fraser, W. R. (2014). Ecological sexual dimorphism and environmental variability within a community of antarctic penguins (genus pygoscelis). PloS One, 9(3), e90081.
LTER, P. S. A., & Gorman, K. (2016). Structural size measurements and isotopic signatures of foraging among adult male and female chinstrap penguins (pygoscelis antarctica) nesting along the palmer archipelago near palmer station, 2007-2009.
- Posted on:
- January 1, 0001
- Length:
- 5 minute read, 882 words
- See Also: